
This is not an official NIH site and does not represent NIH or US government views.
For official views, visit the NIH NIEHS website.

Publications
2025
-
E. Mailler, A. Singh, M. Jarnik, Y. Li, L. Holtzclaw, V. Hoffmann, S. Mine, P. Stallcup, L. Ordoubadinia, and C.M. Guardia^. Liver-specific loss of Atg9a perturbs lipid metabolism and hepatocyte integrity. Autophagy Reports (Accepted).
-
C.E. Kendal-Wright, J.J. Moore, L. Feng, R. Menon, A. L. David, T.T. Chowdhury, M.F. Crowther, C. Howe, N. Zhong, V.Z. Clavellina, P. Flores-Espinosa, N. Truong, V. Sapin-Deour, L. Blanchon, A. Zahra, A.K. Kammala, R. Cherukuri, L.S. Richardson^, Fetal Membrane Society Consortium. A call to standardize the nomenclature of human fetal membrane at the feo-maternal interface. Placenta. 170:42-52 (link).
-
S.M. Ahmadi, M.L. Perez, and C.M. Guardia^. Altered Secretion of placental peptide hormones: functions and trafficking. Front. Endocrinol. 16:1584303 (link).
2024
-
A. Singh, M.L. Perez, O. Kirsanov, E. Padilla-Banks, and C.M. Guardia^. Autophagy in reproduction and pregnancy-associated diseases. iScience. 27(12):111268 (link).
-
E. Bayam^, P. Tilly, S.C. Collins, J. Rivera Alvarez, M. Kannan, L. Tonneau, E. Brivio, B. Rinaldi, R. Lecat, N. Schwaller, L. Cotellessa, S. Maddirevula, F. Monteiro, C.M. Guardia, J.P. Kitajima, F. Kok, M. Kato, A.A.A. Hamed, M.A. Salih, S. Al Tala, M.O. Hashem, H. Tada, H. Saitsu, M. Stabile, P. Giacobini, S. Friant, Z. Yüksel, M. Nakashima, F.S. Alkuraya, B. Yalcin*^, and J.D. Godin*^. Bi-allelic variants in WDR47 cause a complex neurodevelopmental syndrome. EMBO Mol Med. 2024 (link).
-
B.D. Nturubika^, C.M. Guardia, D.C. Gershlick, J.M. Logan, C. Martini, J.K. Heatlie, J.Lazniewska, C. Moore, G.T. Lam, K.L. Li, B.S-Y. Ung, R.D. Brooks, S.M. Hickey, A.G. Bert, P.A. Gregory, L.M. Butler, J.J. O’Leary, D.A. Brooks^*, and I.R.D. Johnson*. Altered expression of vesicular trafficking machinery in prostate cancer affects lysosomal dynamics and provides insight into the underlying biology and disease progression. Br. J. Cancer 2024 (link).
-
P.M. Couto, C.M. Guardia, F.L. Couto, C.A. Labriola, M.S. Labanda, and J.J. Caramelo^. Acceptors stability modulates the efficiency of post-translational protein N-glycosylation. FASEB J. 38:e23782 (link).
2022
-
M. Ishida, M.G. Otero, C. Freeman, P.A. Sánchez-Lara, C.M. Guardia, T.M. Pierson^ & J.S. Bonifacino^. A neurodevelopmental disorder associated with an activating de novo missense variant in ARF1. Hum Mol Genet. ddac279 (link)
-
G. Hu*, P.J. Hauk*, N. Zhang, W. Elsegeiny, C.M. Guardia, A. Kullas, K. Crosby, R.R. Deterding, M. Schedel, P. Reynolds, J.K. Abbott, V. Knight, S. Pittaluga, M. Raffeld, S.D. Rosenzweig, J.S. Bonifacino, G. Uzel, P.R. Williamson^, and E.W. Gelfand. Autophagy-associated immune dysregulation and hyperplasia in a patient with compound heterozygous mutations in ATG9A. Autophagy (link).
-
C.D. Williamson, C.M. Guardia, R. De Pace, J.S. Bonifacino, and A. Saric^. Measurement of lysosome positioning by shell analysis and line scan. Methods Mol Biol. 2473:285-306 (link).
-
T. Ahmad, D. Vullhorst, R. Chaudhuri, C.M. Guardia, N. Chaudhary, I. Karavanova, J.S. Bonifacino & A. Buonanno^. Transcytosis and trans-synaptic retention by postsynaptic ErbB4 underlie axonal accumulation of NRG3. J Cell Biol. 221(7):e202110167 (link)
2021
-
E. Mailler, C.M. Guardia, X. Bai, M. Jarnik, C.D. Williamson, Y. Li, N. Maio, A. Golden, & J.S. Bonifacino^. The autophagy protein ATG9A enables lipid mobilization from lipid droplets. Nat Commun. 12, 6750 (link)
-
C.M. Guardia, E. Kane, A.G. Tebo, A. Sanders, D. Kaya, & K.E. Grogan^. The power of peer networking for improving faculty job applications: a successful pilot program. Submitted (BioRXiv - DOI: 10.1101/2021.10.16.464662)
-
J.L. Nieto-Torres*^, J. Durgan*, A. Franco-Romero*, P. Grumati*, C.M. Guardia*, A.M. Leidal*, M.A. Mandell*, C.G. Towers*, & F. Wang*. The Autophagy, Inflammation and Metabolism Center international eSymposium – an early-career investigators’ seminar series during the COVID-19 pandemic. J Cell Sci 134 (19): jcs259268 (link)
-
C.M. Guardia^, A. Hierro, & D.C. Gershlick^. Editorial: Energy Requirements in Membrane Trafficking. Front Cell Dev Biol. 9:750633 (link)
-
A. Saric, S. Freeman, C.D. Williamson, M. Jarnik, C.M. Guardia, M. Fernandopulle, D.C. Gershlick, & J.S. Bonifacino^. SNX19 restricts endolysosome motility through contacts with the endoplasmic reticulum. Nat Commun. 12, 4552 (link)
-
C.M. Guardia, A. Jain, R. Mattera, A. Friefeld, Y. Li, & J.S. Bonifacino^. RUSC2 and WDR47 oppositely regulate kinesin-1-dependent distribution of ATG9A to the cell periphery. Mol Biol Cell. mbcE21060295 (link)
-
D.J. Klionsky^, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy DOI: 10.1080/15548627.2020.1797280
-
A.D. Senol*, M. Samarani*, S. Syan, C.M. Guardia, T. Nonaka, N. Liv, P.L. Lambert, M. Hasegawa, J. Klumperman, J.S. Bonifacino, & C. Zurzolo^. α-Syn fibrils affect lysosome structure & function to propagate misfolding through TNTs. PLoS Biol 19(7): e3001287 (link)
2020
-
C.M. Guardia*, E.T. Christenson*, W. Zhou, X. Tan, T. Lian, J.D. Faraldo-Gómez, J.S. Bonifacino, J. Jiang & A. Banerjee^. The structure of human ATG9A and its interplay with the lipid bilayer. Autophagy DOI: 10.1080/15548627.2020.1830522
-
C.M. Guardia*, X. Tan*, T. Lian*, M.S. Rana*, W. Zhou, E.T. Christenson, A.J. Lowry, J.D. Faraldo-Gómez, J.S. Bonifacino^, J. Jiang^, & A. Banerjee^. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Reports 31(13), 107837 (link)
-
Editor’s Pick: W.D. Hawkins & D.J. Klionsky. Structure of Human ATG9A: How holey art thou? Autophagy DOI: 10.1080/15548627.2020.1810901
-
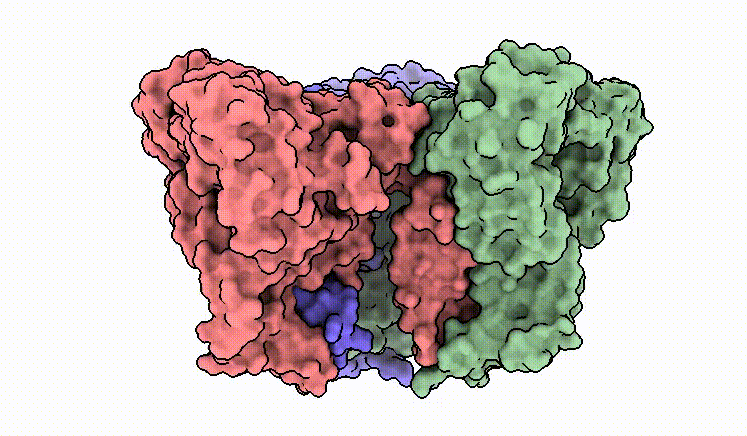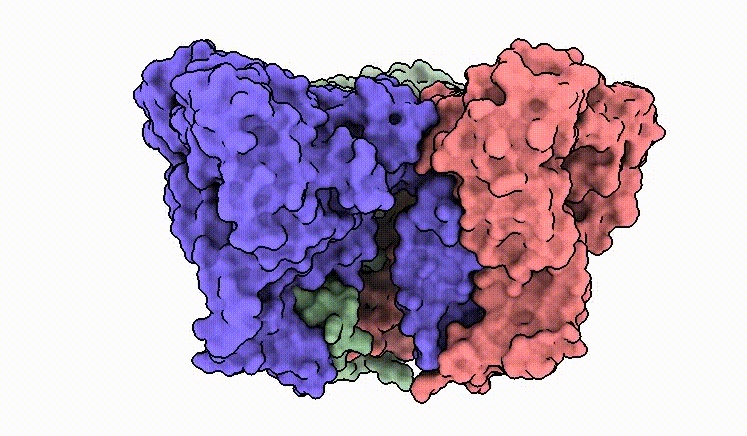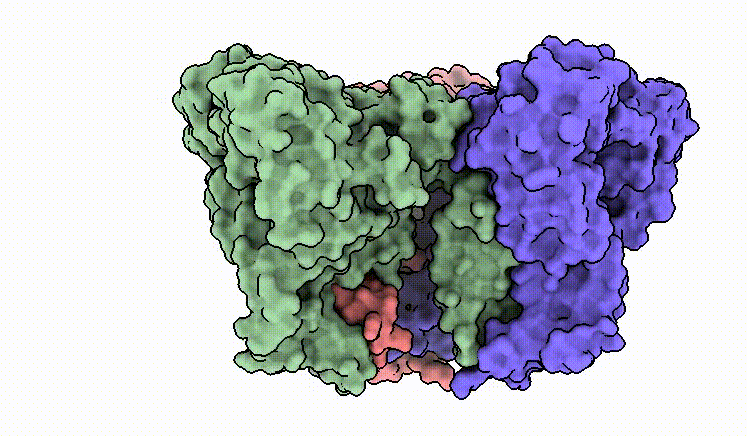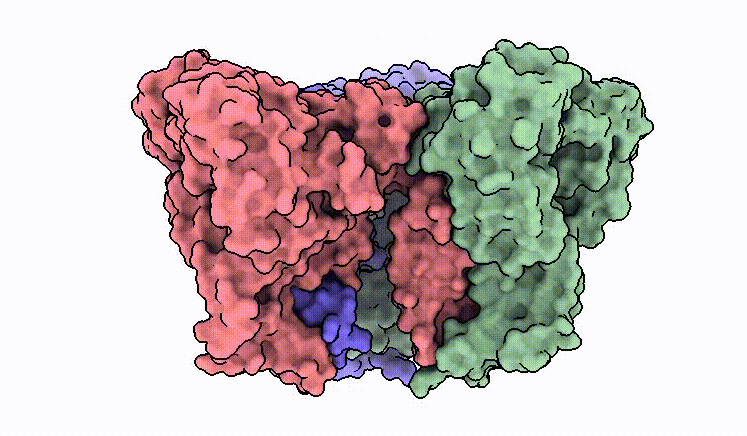
Our cryo-EM structure of human ATG9A
2019
-
C.M. Guardia, R. DePace, A. Sen, A. Saric, D.A. Kolin, A. Kunwar, & J.S. Bonifacino^. Reversible association with motor proteins (RAMP): a streptavidin-based dimerization method to manipulate organelle positioning. PLoS Biol. 17(5): e3000279. (link)
-
S. Gaurav*, C.M. Guardia*, A. Roy, A. Vassilev, A. Saric, L.N. Griner, J. Marugan, J. Marugan, M. Ferrer, J.S. Bonifacino, & M.L. DePamphilis^. A family of PIKfyve inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupts multiple events in lysosome homeostasis. Autophagy, 26:1-25. (link)
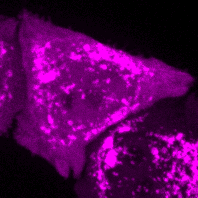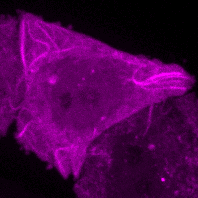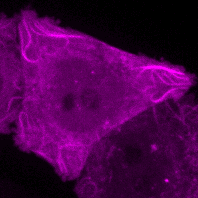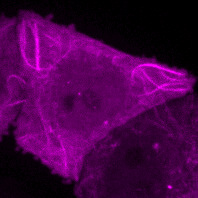
Manipulating organelles
inside the cell with RAMP
2018
-
C.M. Guardia, R. DePace, R. Mattera, & J.S. Bonifacino^. (Review). Neuronal functions of adaptor complexes involved in protein sorting. Curr Opin Neurobiol, 51, 103-10. (link)
2017
-
R. Mattera*, S. Yoon Park*, R. DePace, C.M. Guardia, & J.S. Bonifacino^. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. PNAS, 114(50), E10697-706. (link)
-
R. Jia, C.M. Guardia, J. Pu, Y. Chen & J.S. Bonifacino^. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy, 13(10):1648-63. (link)
-
G.G. Farías, C.M. Guardia, R. De Pace, D.J. Britt & J.S. Bonifacino^. BORC/Kinesin-1 ensemble drives polarized transport of lysosomes into the axon. PNAS, 114(14), E2955-64. (link)
2016
-
J. Pu, C.M. Guardia, T. Keren-Kaplan & J.S. Bonifacino^. (Review). Mechanisms and functions of lysosome positioning. Journal of Cell Science, 0, 1-11. (link)
-
C.M. Guardia, G.G. Farías, J. Pu, R. Jia & J.S. Bonifacino^. BORC functions upstream of kinesins 1 and 3 to regulate regional movement of lysosomes along different microtubule tracks. Cell Reports, 17(8), 1950-61. (link)
-
D.J. Britt, G.G. Farías, C.M. Guardia, & J.S. Bonifacino^. (Review). Mechanisms of polarized organelle distribution in neurons. Front. Cell Neurosci. 10(88), 7842-57. (link)
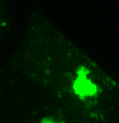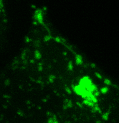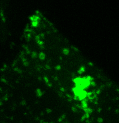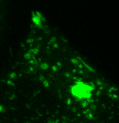
Lysosomes moving and stretching
inside the cell with the help of kinesins
2015
-
G.G. Farías, C.M. Guardia, D.J. Britt, X. Guo, & J.S. Bonifacino^. Sorting of dendritic and axonal vesicles at the pre-axonal exclusion zone. Cell Reports 13(6), 1221-32. (link)
-
R. Mattera, C.M. Guardia, S.S. Sidhu & J.S. Bonifacino^. Bivalent motif-ear interactions mediate the association of the accessory protein tepsin with the AP-4 adaptor complex. J. Biol. Chem. 290(52), 30736-49. (link)
2014
-
C.M. Guardia, J.J. Caramelo, M. Trujillo, S.P. M-Huergo, R. Radi, D.A. Estrin & G.A. Rabinovich^. Structural basis of redox-dependent modulation of galectin-1 dynamics and function. Glycobiology 24(5), 428-41. (link)
-
A. Zeida, C.M. Guardia, P. Lichtig, L.L. Perissinotti, L.A. Defelipe, A. Turjanski, R. Radi, M. Trujillo & D.A. Estrin^. (Review). Thiol redox biochemistry: insights from computer simulations. Biophysical Reviews 1(6), 27-46. (link)
2011
-
S. Di Lella, V. Sundblad, J.P. Cerliani, C.M. Guardia, D.A. Estrin, G.R. Vasta & G.A. Rabinovich^. (Review). When galectins recognize glycans: from biochemistry to physiology and back again. Biochemistry 50(37), 7842-57. (link)
-
C.M.A. Guardia, D. Gauto, S. Di Lella, G. Rabinovich, M.A. Martí & D.A. Estrin^. An integrated computational analysis of structure, dynamics and ligand binding interaction of the human galectin network. J. Chem. Inf. Model. 51(8), 1918-30. Cover of the Issue (link)
2010
-
S. Di Lella, M.A. Marti, D. Croci, C.M.A. Guardia, J. Diaz Ricci, G.A. Rabinovich, J.J. Caramelo & D.A. Estrin^. Linking structure and thermal stability of the beta-galactosidase binding protein galectin-1 to ligand binding and dimerization equilibria. Biochemistry 49(35), 7652-58. (link)
2009
-
D.F. Gauto, S. Di Lella, C.M.A. Guardia, D.A. Estrin & M.A. Martí^. Carbohydrate-binding proteins: dissecting ligand structures through solvent environment occupancy. J. Phys. Chem. B 113(25): 8717-24. (link)
2008
-
C.M.A. Guardia, M.C. González Lebrero, S.E. Bari & D.A. Estrin^. QM/MM investigation of the reaction products between nitroxyl and O2 in aqueous solution. Chem. Phys. Lett. 463, 112-16. (link)
* denotes equal contribution
^ denotes corresponding authors
More information is available at Google Scholar